Protein Name: Transcription factor AP-1
|
UniprotKB/SwissProt ID: JUN_HUMAN (P05412)
Gene Name: JUN
Organism: Homo sapiens (Human).
Function: Transcription factor that recognizes and binds to the enhancer heptamer motif 5'-TGA[CG]TCA-3'. Promotes activity of NR5A1 when phosphorylated by HIPK3 leading to increased steroidogenic gene expression upon cAMP signaling pathway stimulation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00020 | Colorectal cancer |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 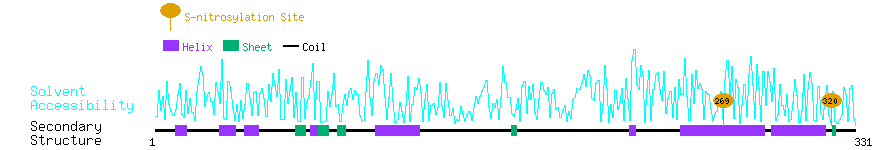 |
|

