Protein Name: Calcium/calmodulin-dependent protein kinase type II subunit gamma
|
UniprotKB/SwissProt ID: KCC2G_HUMAN (Q13555)
Gene Name: CAMK2G
Synonyms: CAMK, CAMK-II, CAMKG
Organism: Homo sapiens (Human).
Function: Calcium/calmodulin-dependent protein kinase that functions autonomously after Ca(2+)/calmodulin-binding and autophosphorylation, and is involved in sarcoplsamic reticulum Ca(2+) transport in skeletal muscle and may function in dendritic spine and synapse formation and neuronal plasticity. In slow- twitch muscles, is involved in regulation of sarcoplasmic reticulum (SR) Ca(2+) transport and in fast-twitch muscle participates in the control of Ca(2+) release from the SR through phosphorylation of the ryanodine receptor-coupling factor triadin. In neurons, may participate in the promotion of dendritic spine and synapse formation and maintenance of synaptic plasticity which enables long-term potentiation (LTP) and hippocampus-dependent learning.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Sarcoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side (Probable).
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 602123 | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE II-GAMMA; CAMK2G ;;CaM KINASE II GAMMA SUBUNIT;; |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 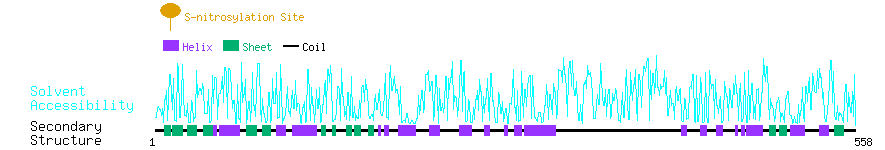 |
|

