Protein Name: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
|
UniprotKB/SwissProt ID: NDUS7_MOUSE (Q9DC70)
Gene Name: Ndufs7
Organism: Mus musculus (Mouse).
Function: Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 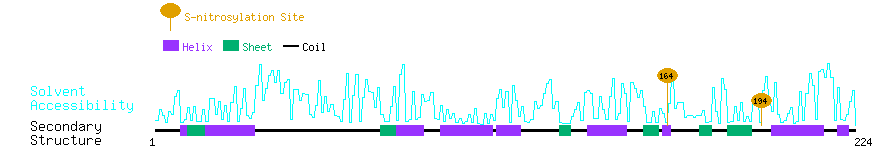 |
|

