Protein Name: Omega-amidase NIT2
|
UniprotKB/SwissProt ID: NIT2_HUMAN (Q9NQR4)
Gene Name: NIT2
Organism: Homo sapiens (Human).
Function: Has a omega-amidase activity. The role of omega-amidase is to remove potentially toxic intermediates by converting alpha- ketoglutaramate and alpha-ketosuccinamate to biologically useful alpha-ketoglutarate and oxaloacetate, respectively. Overexpression decreases the colony-forming capacity of cultured cells by arresting cells in the G2 phase of the cell cycle.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 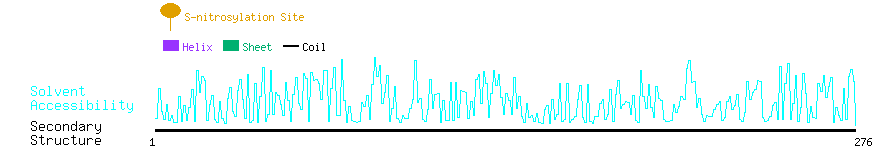 |
|

