Protein Name: Vesicle-fusing ATPase
|
UniprotKB/SwissProt ID: NSF_HUMAN (P46459)
Gene Name: NSF
Organism: Homo sapiens (Human).
Function: Required for vesicle-mediated transport. Catalyzes the fusion of transport vesicles within the Golgi cisternae. Is also required for transport from the endoplasmic reticulum to the Golgi stack. Seems to function as a fusion protein required for the delivery of cargo proteins to all compartments of the Golgi stack independent of vesicle origin. Interaction with AMPAR subunit GRIA2 leads to influence GRIA2 membrane cycling (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 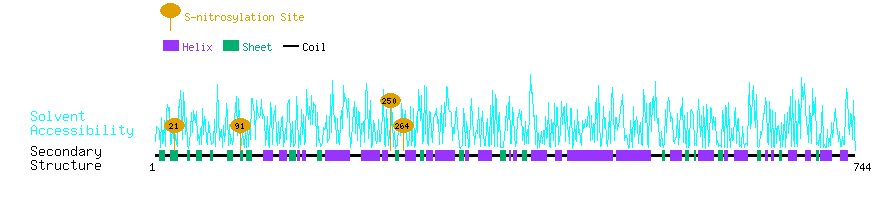 |
|

