Protein Name: 2-oxoglutarate dehydrogenase, mitochondrial
|
UniprotKB/SwissProt ID: ODO1_RAT (Q5XI78)
Gene Name: Ogdh
Synonyms: Ogdhl
Organism: Rattus norvegicus (Rat).
Function: The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components: 2- oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3) (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 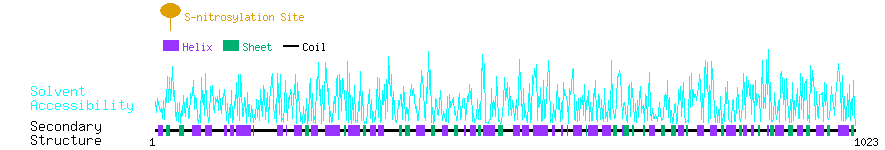 |
|

