Protein Name: Prostaglandin E synthase 2
|
UniprotKB/SwissProt ID: PGES2_MOUSE (Q8BWM0)
Gene Name: Ptges2
Synonyms: Gbf1, Pges2
Organism: Mus musculus (Mouse).
Function: Dual function enzyme which exhibits both isomerase and lyase activities. Its catalytic activity is altered by the presence or absence of the cofactors glutathione (GSH) and heme. The GSH-heme complex-bound enzyme (mPGES-2h) acts as a lyase and catalyzes the degradation of prostaglandin E2 H2 (PGH2) to 12(S)- hydroxy-5(Z),8(E),10(E)-heptadecatrienoic acid (HHT) and malondialdehyde (MDA) while the GSH-heme complex-free enzyme (mPGES-2) acts as an isomerase and catalyzes the isomerization of PGH2 into the more stable prostaglandin E2 (PGE2) form (By similarity). May also have transactivation activity toward IFN- gamma (IFNG), possibly via an interaction with CEBPB; however, the relevance of transcription activation activity remains unclear.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Golgi apparatus membrane; Single-pass membrane protein. Nucleus. Note=According to PubMed:12050152, some fraction may be nuclear. Prostaglandin E synthase 2 truncated form: Cytoplasm. Note=Synthesized as a Golgi membrane-bound protein, which is further c
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 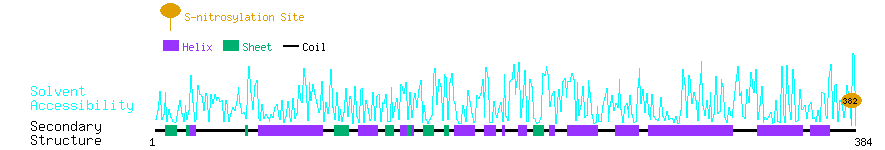 |
|

