Protein Name: Phosphoglycolate phosphatase
|
UniprotKB/SwissProt ID: PGP_MOUSE (Q8CHP8)
Gene Name: Pgp
Organism: Mus musculus (Mouse).
Function: Phosphatase with broad substrate specificity. Has high phosphatase activity toward ADP, ATP, GDP, GTP and p- nitrophenylphosphate. Has tyrosine-protein phosphatase activity (in vitro). Has very low activity with pyridoxal 5'-phosphate.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 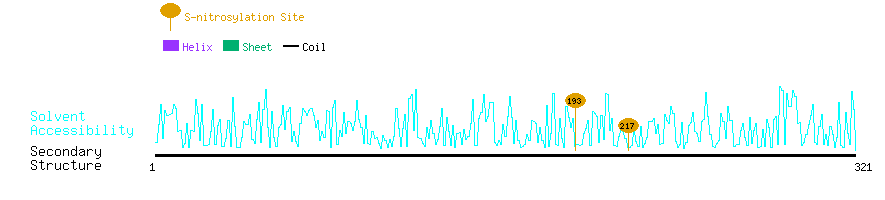 |
|
