Protein Name: 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma
|
UniprotKB/SwissProt ID: PLCC_MOUSE (Q9D517)
Gene Name: Agpat3
Organism: Mus musculus (Mouse).
Function: Converts lysophosphatidic acid (LPA) into phosphatidic acid by incorporating an acyl moiety at the sn-2 position of the glycerol backbone. Acts on LPA containing saturated or unsaturated fatty acids C16:0-C20:4 at the sn-1 position using C18:1, C20:4 or C18:2-CoA as the acyl donor. Also acts on lysophosphatidylcholine, lysophosphatidylinositol and lysophosphatidylserine using C18:1 or C20:4-CoA.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Endoplasmic reticulum membrane; Multi-pass membrane protein (By similarity). Nucleus envelope (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 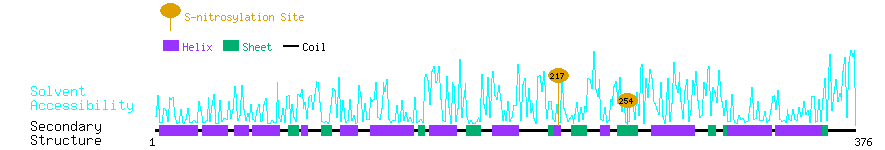 |
|

