Protein Name: Peroxiredoxin-5, mitochondrial
|
UniprotKB/SwissProt ID: PRDX5_HUMAN (P30044)
Gene Name: PRDX5
Synonyms: ACR1
Organism: Homo sapiens (Human).
Function: Reduces hydrogen peroxide and alkyl hydroperoxides with reducing equivalents provided through the thioredoxin system. Involved in intracellular redox signaling.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Isoform Mitochondrial: Mitochondrion. Isoform Cytoplasmic+peroxisomal: Cytoplasm. Peroxisome.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 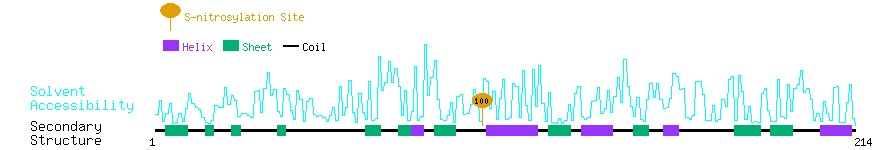 |
|
