Protein Name: 26S proteasome non-ATPase regulatory subunit 11
|
UniprotKB/SwissProt ID: PSD11_HUMAN (O00231)
Gene Name: PSMD11
Organism: Homo sapiens (Human).
Function: Component of the lid subcomplex of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. In the complex, PSMD11 is required for proteasome assembly. Plays a key role in increased proteasome activity in embryonic stem cells (ESCs): its high expression in ESCs promotes enhanced assembly of the 26S proteasome, followed by higher proteasome activity.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus (By similarity). Cytoplasm, cytosol (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 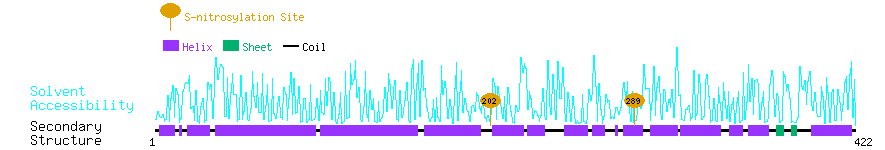 |
|

