Protein Name: 26S proteasome non-ATPase regulatory subunit 1
|
UniprotKB/SwissProt ID: PSMD1_MOUSE (Q3TXS7)
Gene Name: Psmd1
Organism: Mus musculus (Mouse).
Function: Acts as a regulatory subunit of the 26 proteasome which is involved in the ATP-dependent degradation of ubiquitinated proteins (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 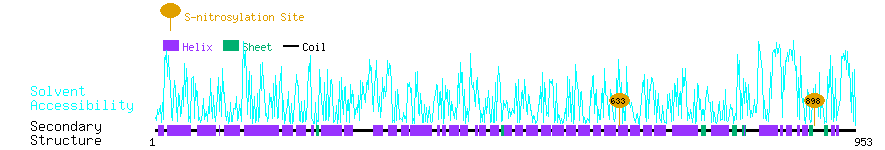 |
|

