Protein Name: Tyrosine-protein phosphatase non-receptor type 1
|
UniprotKB/SwissProt ID: PTN1_MOUSE (P35821)
Gene Name: Ptpn1
Organism: Mus musculus (Mouse).
Function: Tyrosine-protein phosphatase which acts as a regulator of endoplasmic reticulum unfolded protein response. Mediates dephosphorylation of EIF2AK3/PERK; inactivating the protein kinase activity of EIF2AK3/PERK. May play an important role in CKII- and p60c-src-induced signal transduction cascades. May regulate the EFNA5-EPHA3 signaling pathway which modulates cell reorganization and cell-cell repulsion. May also regulate the hepatocyte growth factor receptor signaling pathway through dephosphorylation of MET (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Endoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side (By similarity). Note=Interacts with EPHA3 at the cell membrane (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 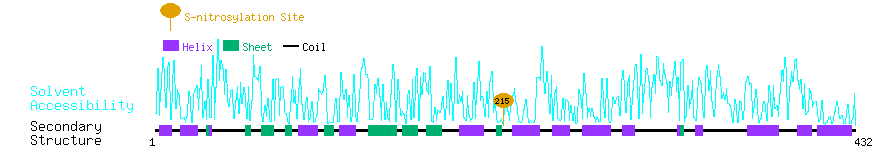 |
|

