Protein Name: GTP-binding nuclear protein Ran
|
UniprotKB/SwissProt ID: RAN_HUMAN (P62826)
Gene Name: RAN
Synonyms: ARA24
Organism: Homo sapiens (Human).
Function: GTP-binding protein involved in nucleocytoplasmic transport. Required for the import of protein into the nucleus and also for RNA export. Involved in chromatin condensation and control of cell cycle (By similarity). The complex with BIRC5/ survivin plays a role in mitotic spindle formation by serving as a physical scaffold to help deliver the RAN effector molecule TPX2 to microtubules. Acts as a negative regulator of the kinase activity of VRK1 and VRK2. Enhances AR-mediated transactivation. Transactivation decreases as the poly-Gln length within AR increases.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus. Cytoplasm. Melanosome. Note=Predominantly nuclear during interphase (By similarity). Becomes dispersed throughout the cytoplasm during mitosis. Identified by mass spectrometry in melanosome fractions from stage I to stage IV.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 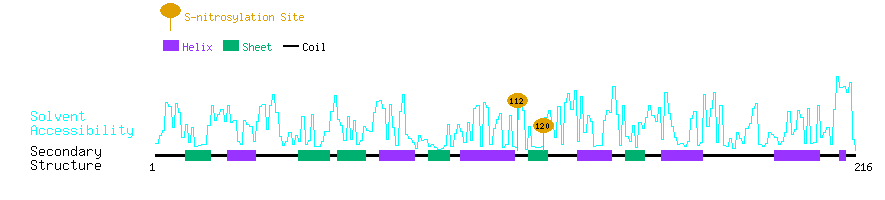 |
|
