Protein Name: GTP-binding protein SAR1a
|
UniprotKB/SwissProt ID: SAR1A_MOUSE (P36536)
Gene Name: Sar1a
Synonyms: Sara, Sara1
Organism: Mus musculus (Mouse).
Function: Involved in transport from the endoplasmic reticulum to the Golgi apparatus. Required to maintain SEC16A localization at discrete locations on the ER membrane perhaps by preventing its dissociation. SAR1A-GTP-dependent assembly of SEC16A on the ER membrane forms an organized scaffold defining endoplasmic reticulum exit sites (ERES) (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Endoplasmic reticulum (By similarity). Golgi apparatus (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 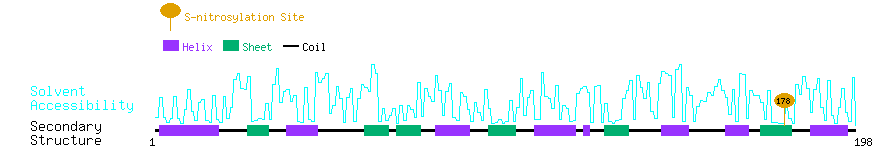 |
|

