Protein Name: Stromal interaction molecule 1
|
UniprotKB/SwissProt ID: STIM1_HUMAN (Q13586)
Gene Name: STIM1
Synonyms: GOK
Organism: Homo sapiens (Human).
Function: Plays a role in mediating store-operated Ca(2+) entry (SOCE), a Ca(2+) influx following depletion of intracellular Ca(2+) stores. Acts as Ca(2+) sensor in the endoplasmic reticulum via its EF-hand domain. Upon Ca(2+) depletion, translocates from the endoplasmic reticulum to the plasma membrane where it activates the Ca(2+) release-activated Ca(2+) (CRAC) channel subunit, TMEM142A/ORAI1.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Single-pass type I membrane protein. Endoplasmic reticulum membrane; Single-pass type I membrane protein. Cytoplasm, cytoskeleton. Note=Translocates from the endoplasmic reticulum to the cell membrane in response to a depletion of intracel
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00093 | Combined immunodeficiencies (CIDs), including the following nine diseases: X-linked hyper IgM syndro | | OMIM | 160565 | MYOPATHY, TUBULAR AGGREGATE ;;TUBULAR AGGREGATE MYOPATHY | | OMIM | 605921 | STROMAL INTERACTION MOLECULE 1; STIM1 | | OMIM | 612783 | IMMUNE DYSFUNCTION WITH T-CELL INACTIVATION DUE TO CALCIUM ENTRY DEFECT 2 |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 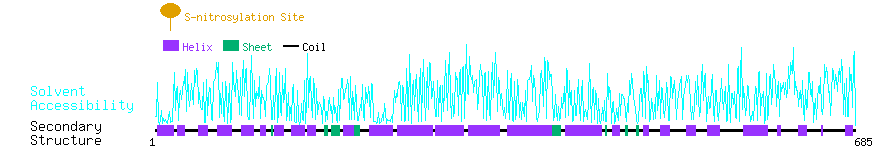 |
|

