Protein Name: Serotransferrin
|
UniprotKB/SwissProt ID: TRFE_HUMAN (P02787)
Gene Name: TF
Organism: Homo sapiens (Human).
Function: Transferrins are iron binding transport proteins which can bind two Fe(3+) ions in association with the binding of an anion, usually bicarbonate. It is responsible for the transport of iron from sites of absorption and heme degradation to those of storage and utilization. Serum transferrin may also have a further role in stimulating cell proliferation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Secreted.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 01811 | Atransferrinemia | | HPRD | 01811 | Iron deficiency anemia, susceptibility to | | HPRD | 01811 | Transferrin variant B2 | | HPRD | 01811 | Transferrin variant BV | | HPRD | 01811 | Transferrin variant C1/C2 | | HPRD | 01811 | Transferrin variant CHI | | HPRD | 01811 | Transferrin variant D1 |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 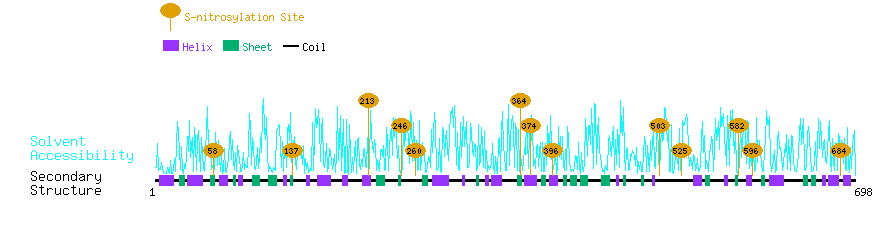 |
|

