Protein Name: Synaptic vesicle membrane protein VAT-1 homolog
|
UniprotKB/SwissProt ID: VAT1_MOUSE (Q62465)
Gene Name: Vat1
Synonyms: Vat-1
Organism: Mus musculus (Mouse).
Function: Plays a part in calcium-regulated keratinocyte activation in epidermal repair mechanisms. Has no effect on cell proliferation (By similarity). Possesses ATPase activity. Negatively regulates mitochondrial fusion in cooperation with mitofusin proteins (MFN1-2) (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Mitochondrion outer membrane; Peripheral membrane protein. Note=The majority is localized in the cytoplasm a small amount is associated with mitochondria (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 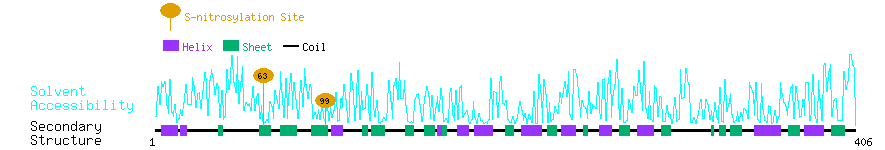 |
|

